Research
Our research tries to answer questions about the evolution of proteins and how this information can be used to create a new range of AI tools.
The tools we have developed make it possible to predict the structure of protein interactions, and to design new molecules that bind to proteins.
We aim to create a universal molecular framework, where any molecule can be predicted.
We are deploying the technology we develop and test it in the lab to create new molecular functions to improve diagnostics and treat diseases such as cancer.
Agonist Design Beyond Natural Amino Acids
By transfer learning from RareFold on high-quality structurtes from GPCRdb, we can learn how to incorporate noncanonical amino acids (NCAAs) seen in RareFold to the structure prediction of GPCRs without ever having seen NCAA-based GPCR modulators. This creates possibilities for new chemistry, immune evasion and degradation resistance.
Li Q, Helleday T and Bryant P. RareFoldGPCR: Agonist Design Beyond Natural Amino Acids. bioRxiv 2025.10.01.679733; doi:10.1101/2025.10.01.679733 Paper Code
Noncanonical Amino Acids
RareFold is a breakthrough deep learning model that accurately predicts protein structures with noncanonical amino acids and enables the design of high-affinity peptide binders (both linear and cyclic) through its EvoBindRare framework. Expanding beyond the 20 standard amino acids, RareFold opens new possibilities for therapeutic design with enhanced stability, specificity, and immune evasion.
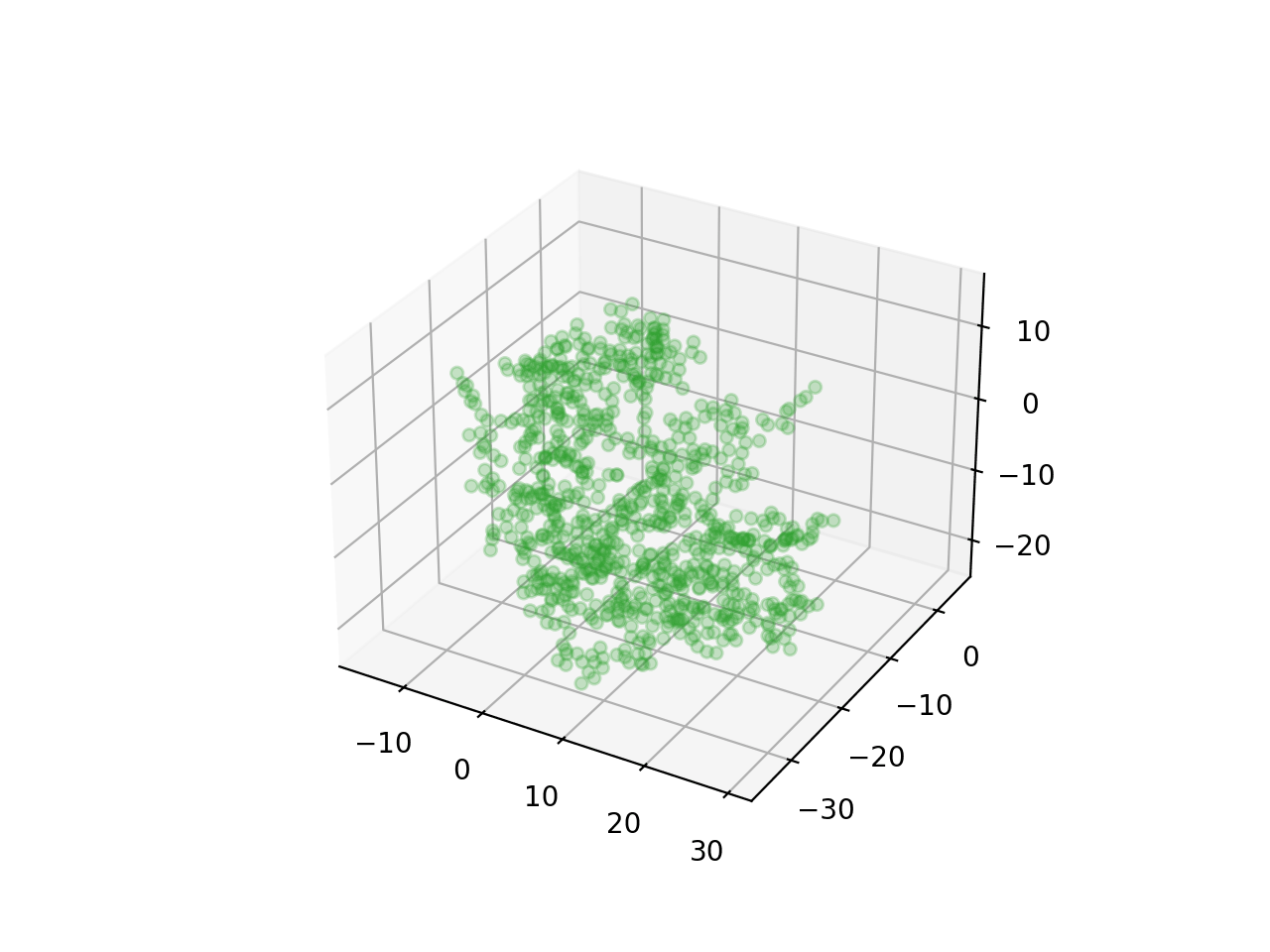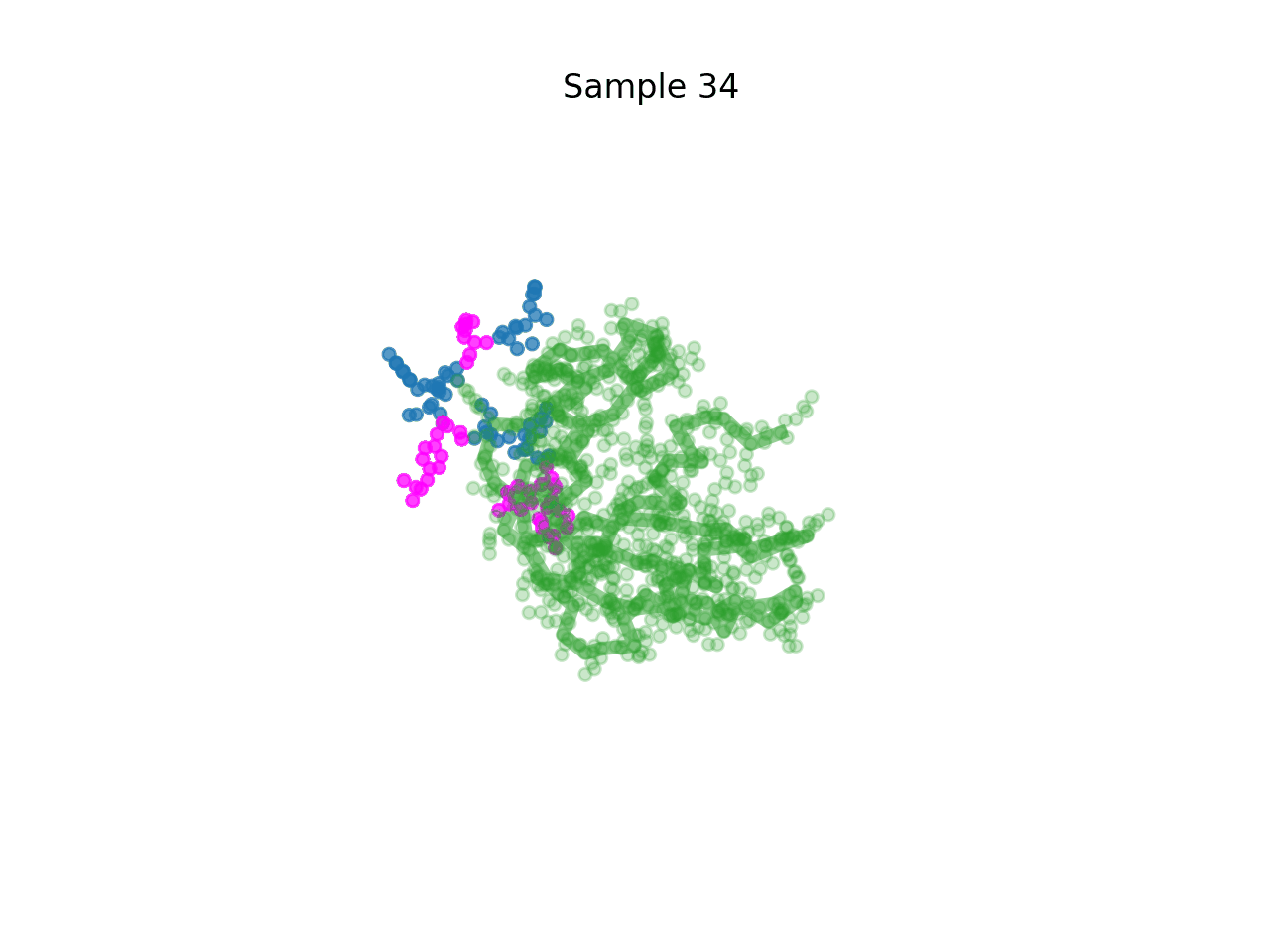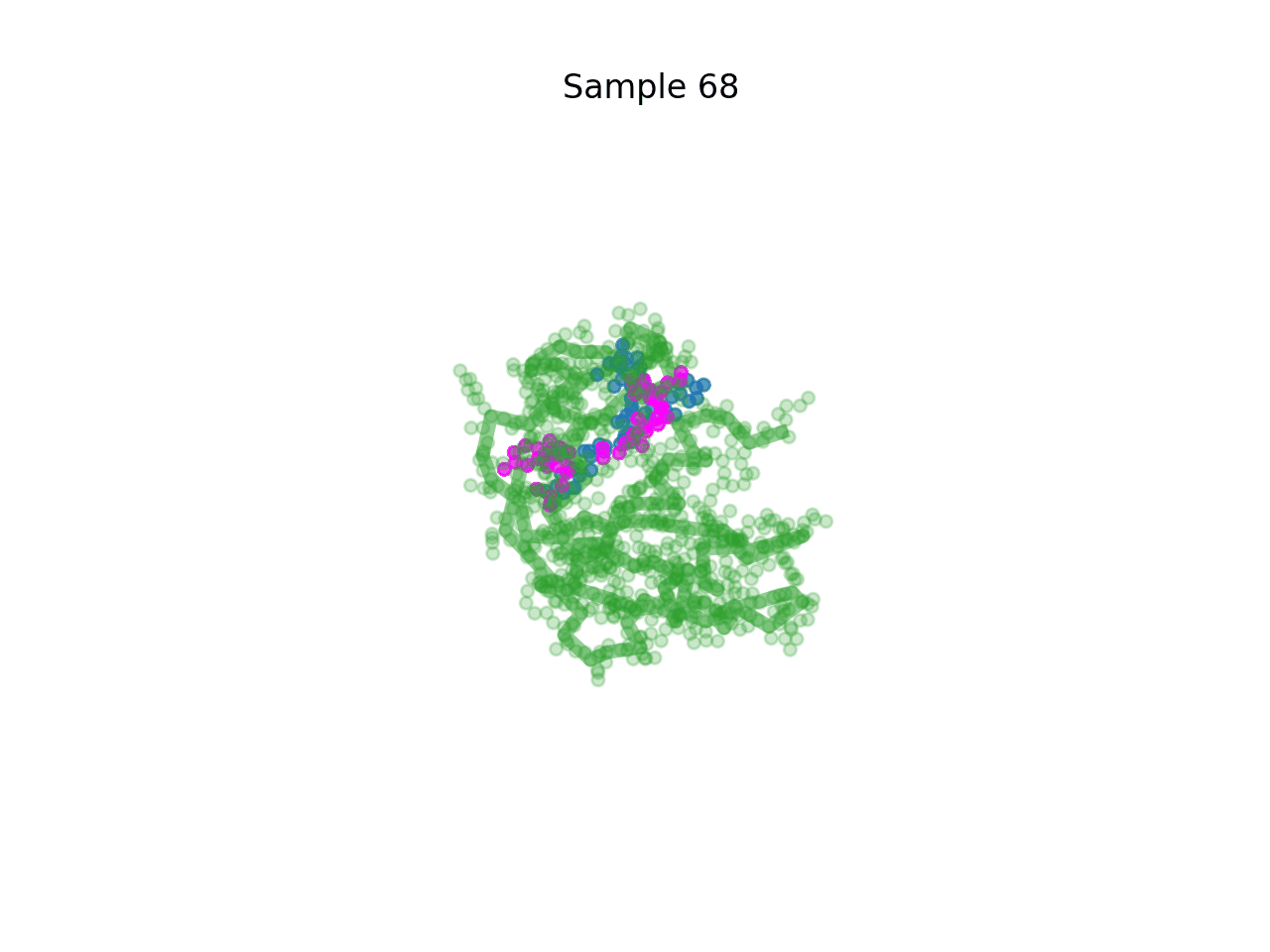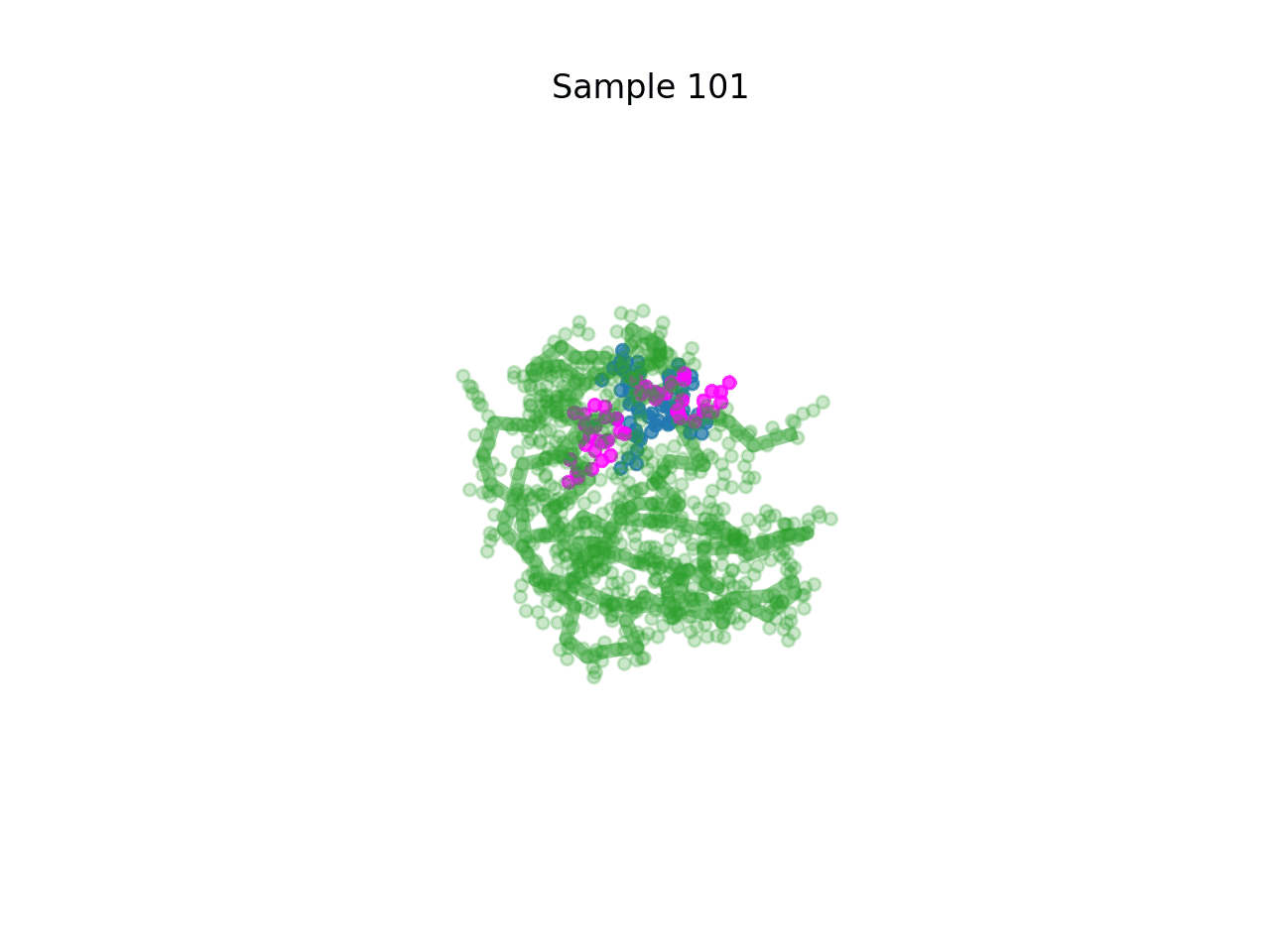
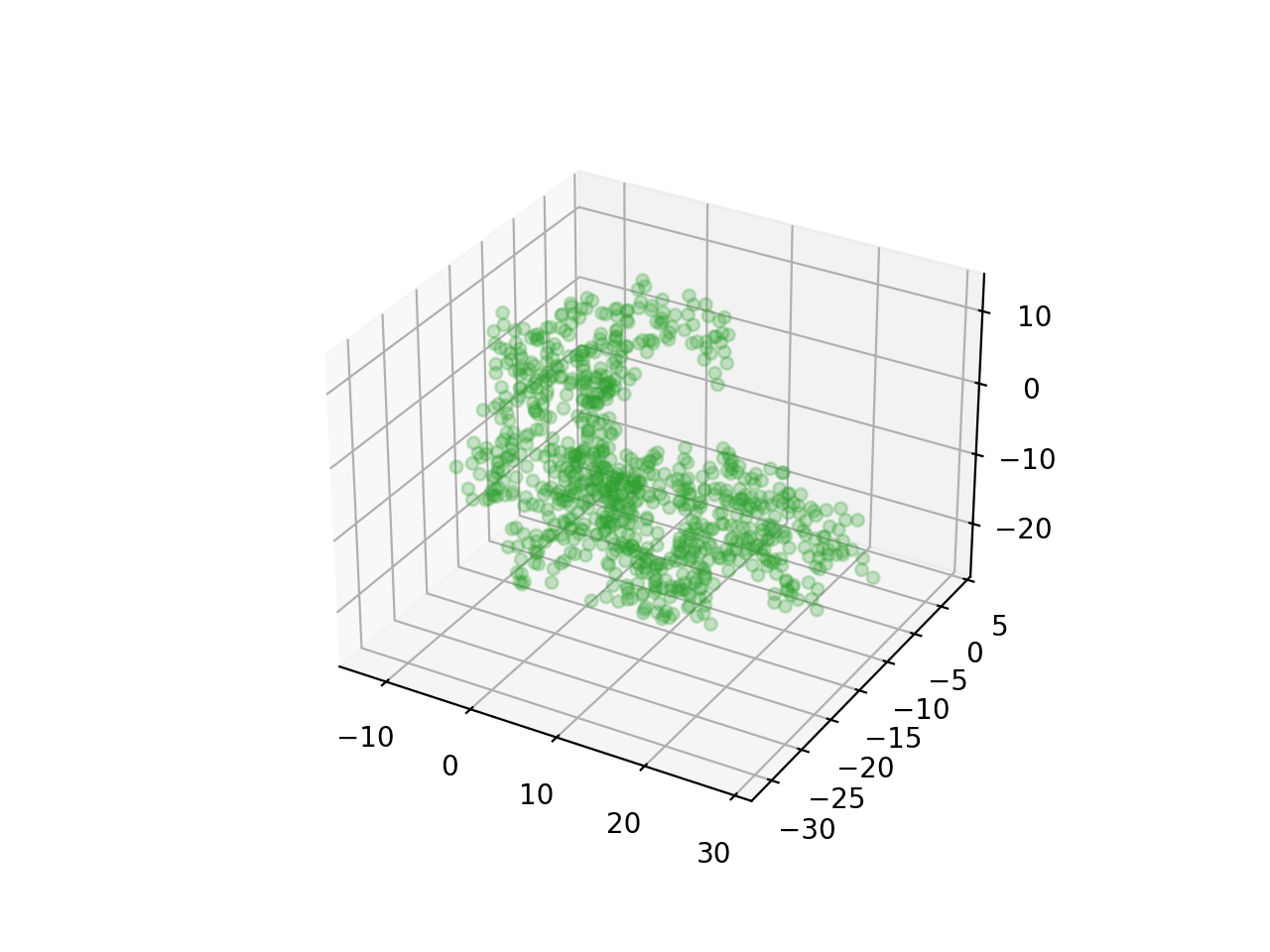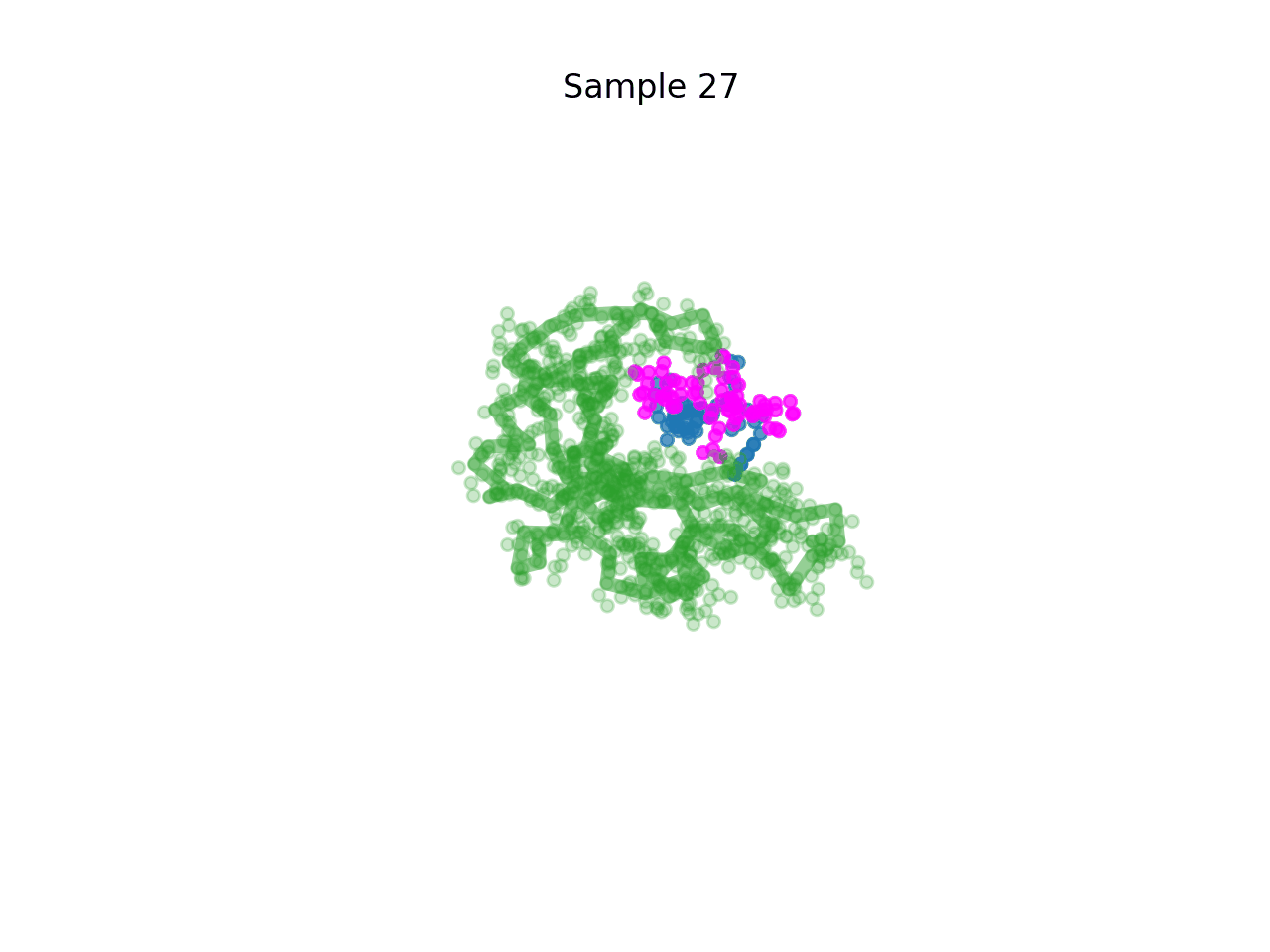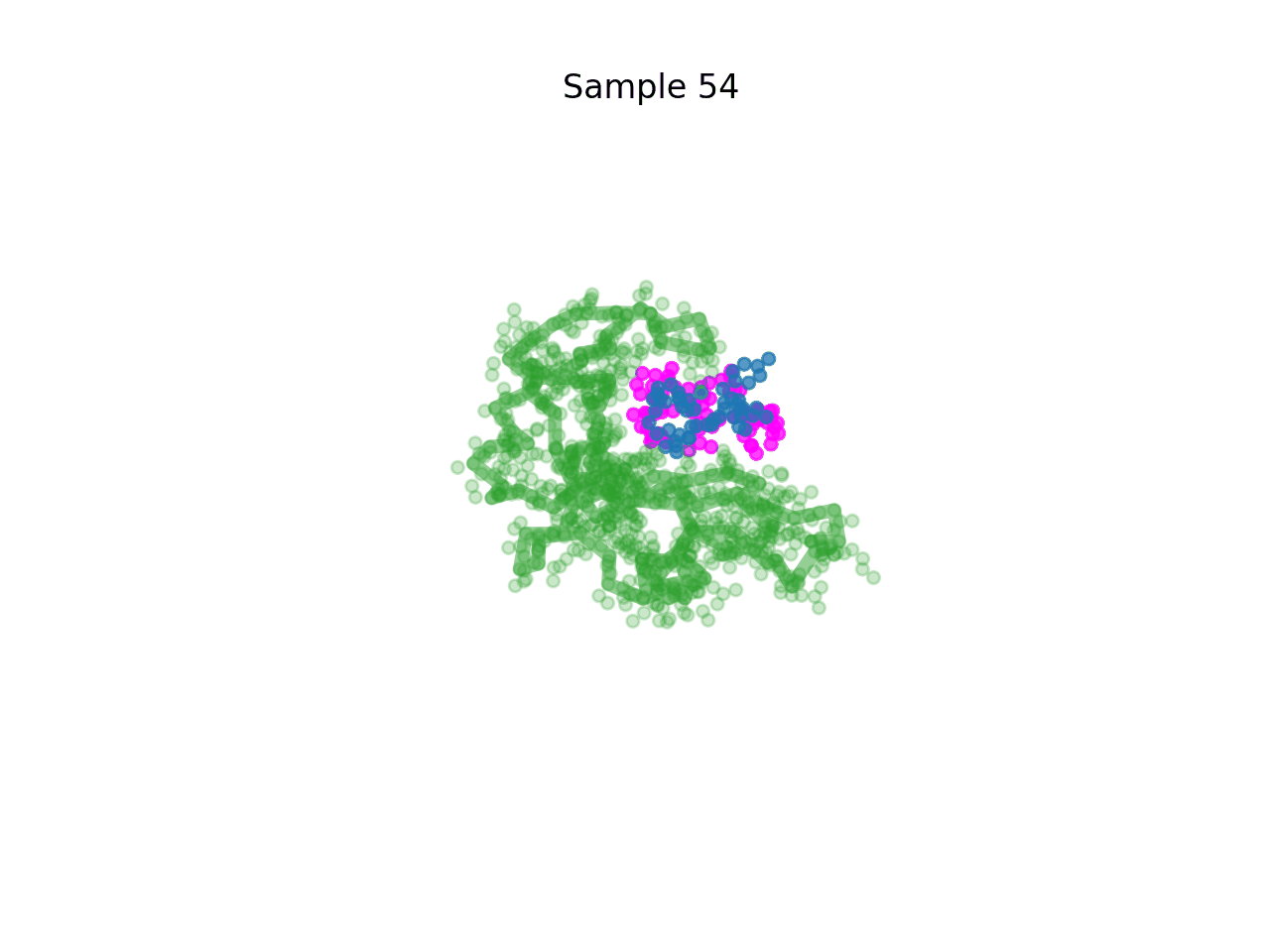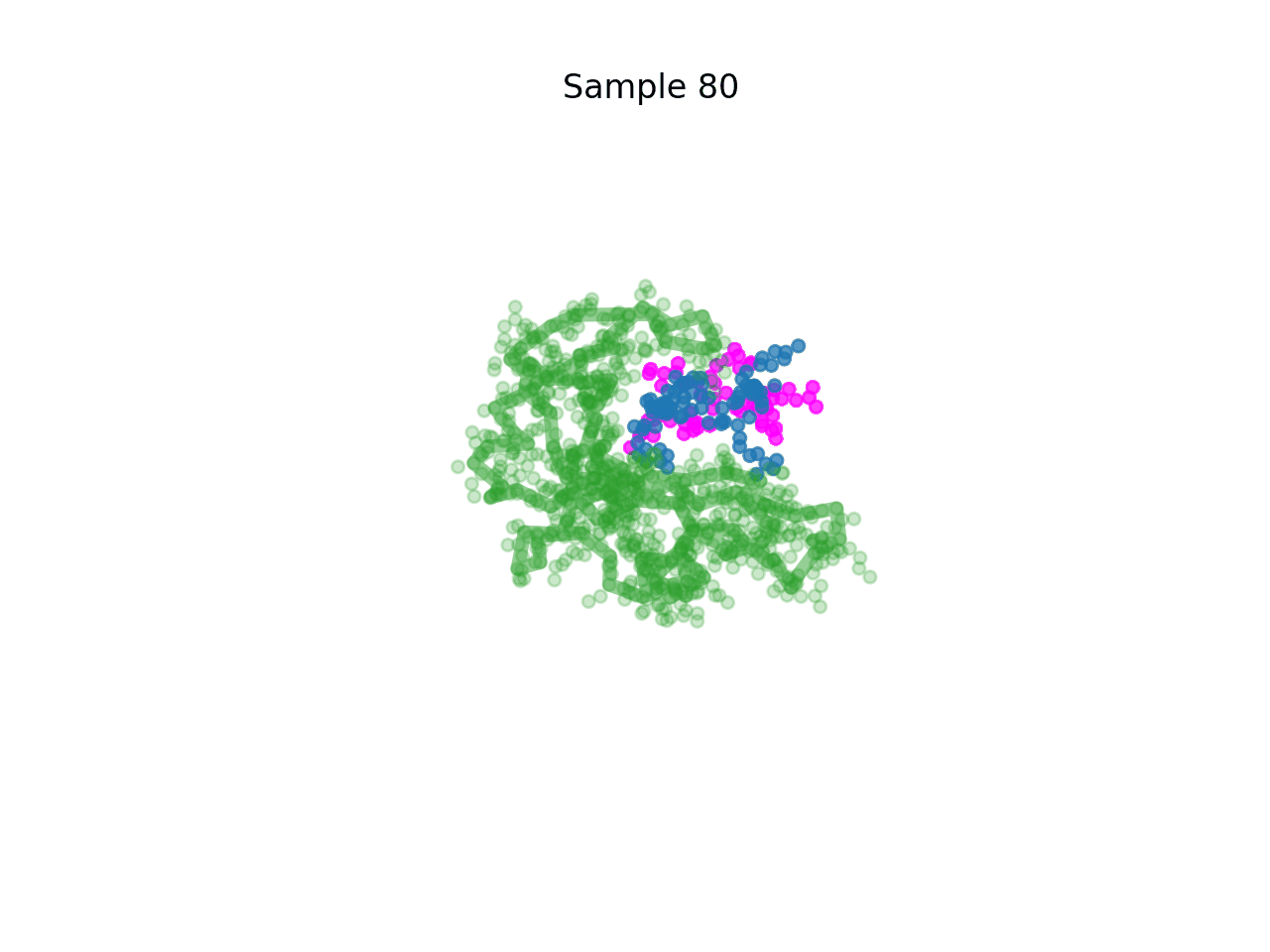
Li Q, Daumiller D, Zuo F, Marcotte H, Pan-Hammarstrom Q and Bryant P. RareFold: Structure prediction and design of proteins with noncanonical amino acids. bioRxiv. 2025. p. 2025.05.19.654846. doi:10.1101/2025.05.19.654846 Paper Code
Agonist design
Li Q, Wiita E, Helleday T, Bryant P. Blind De Novo Design of Dual Cyclic Peptide Agonists Targeting GCGR and GLP1R. bioRxiv. 2025. p. 2025.06.06.658268. doi: https://doi.org/10.1101/2025.06.06.658268 Paper Code
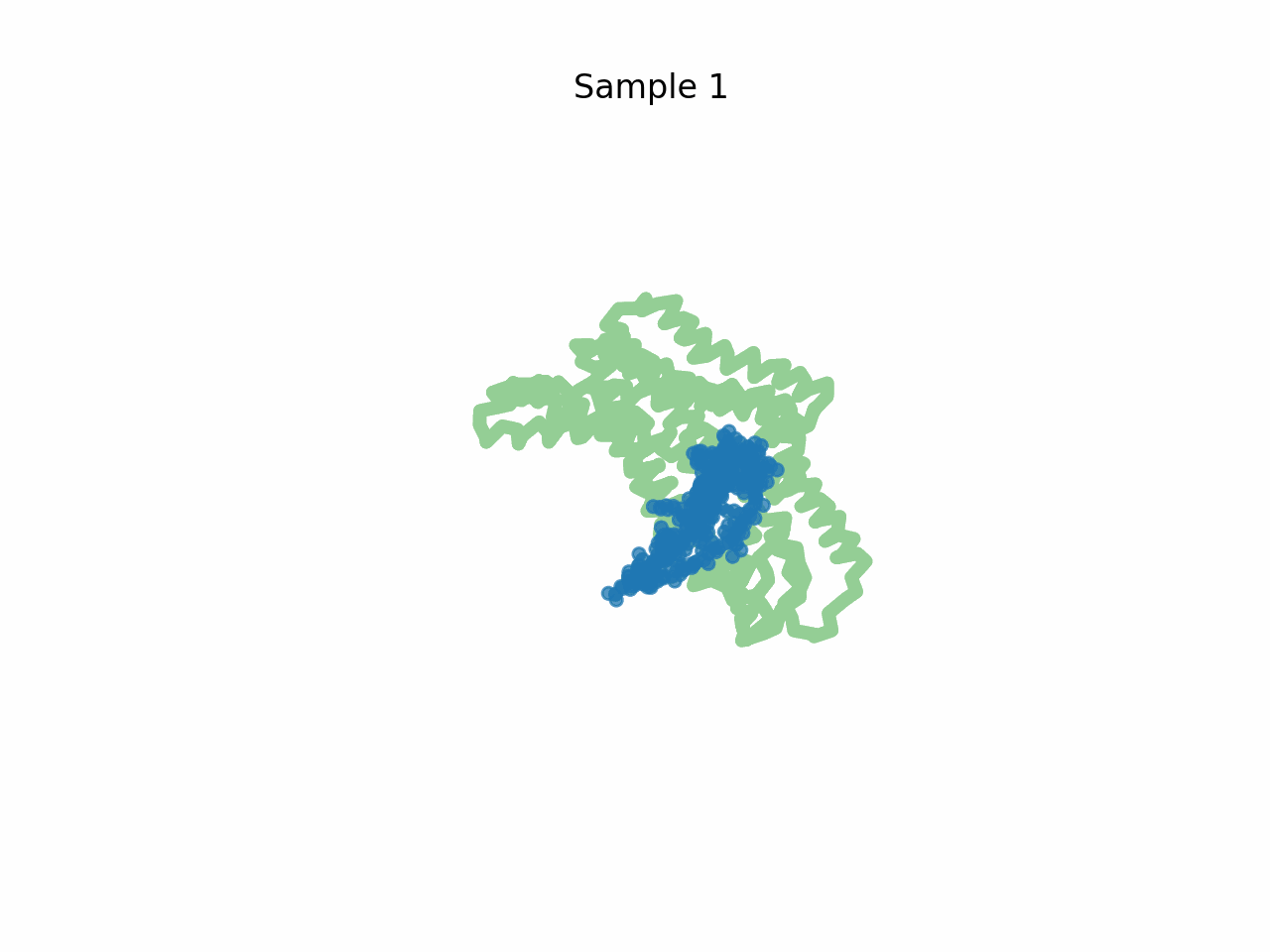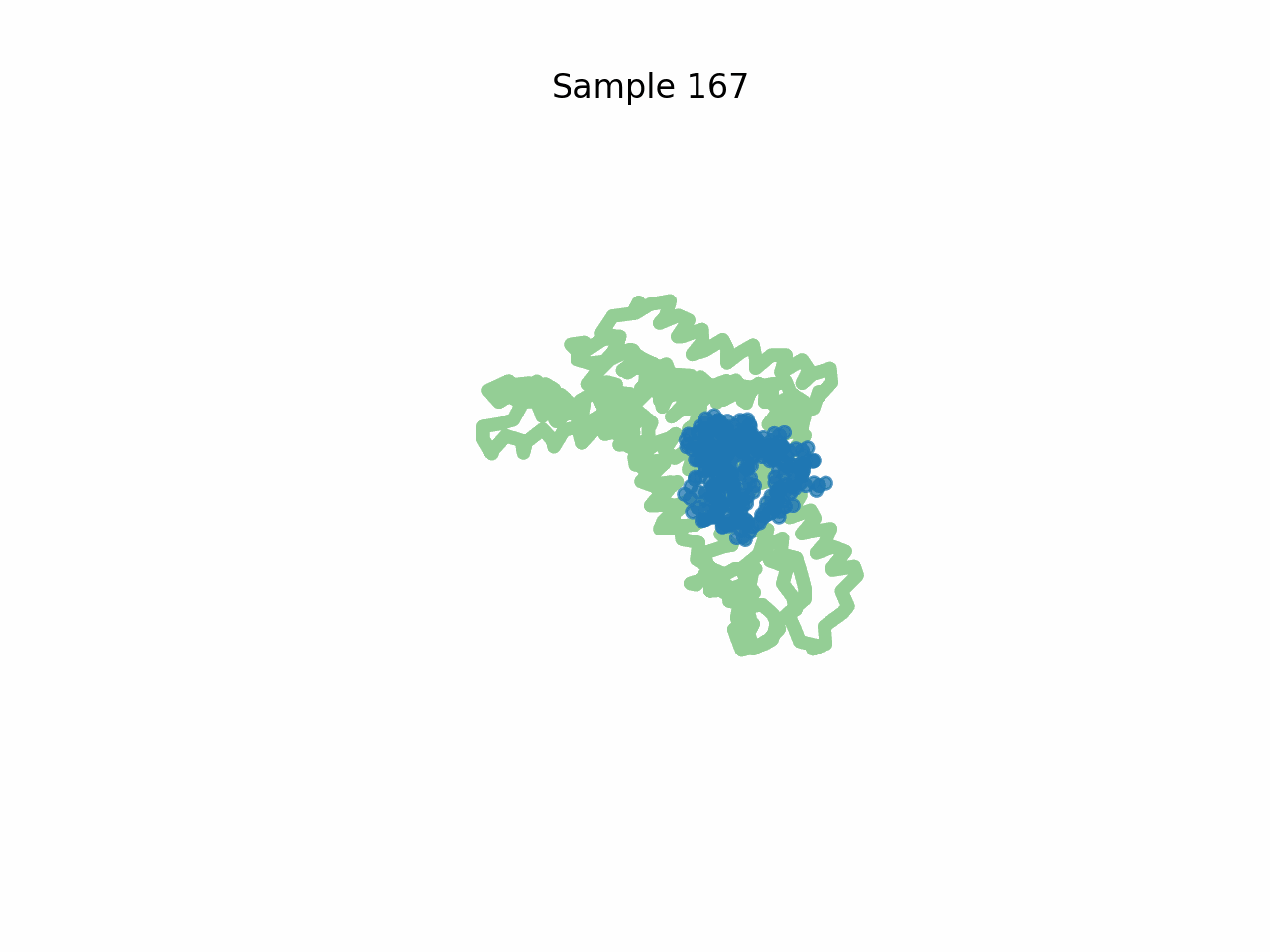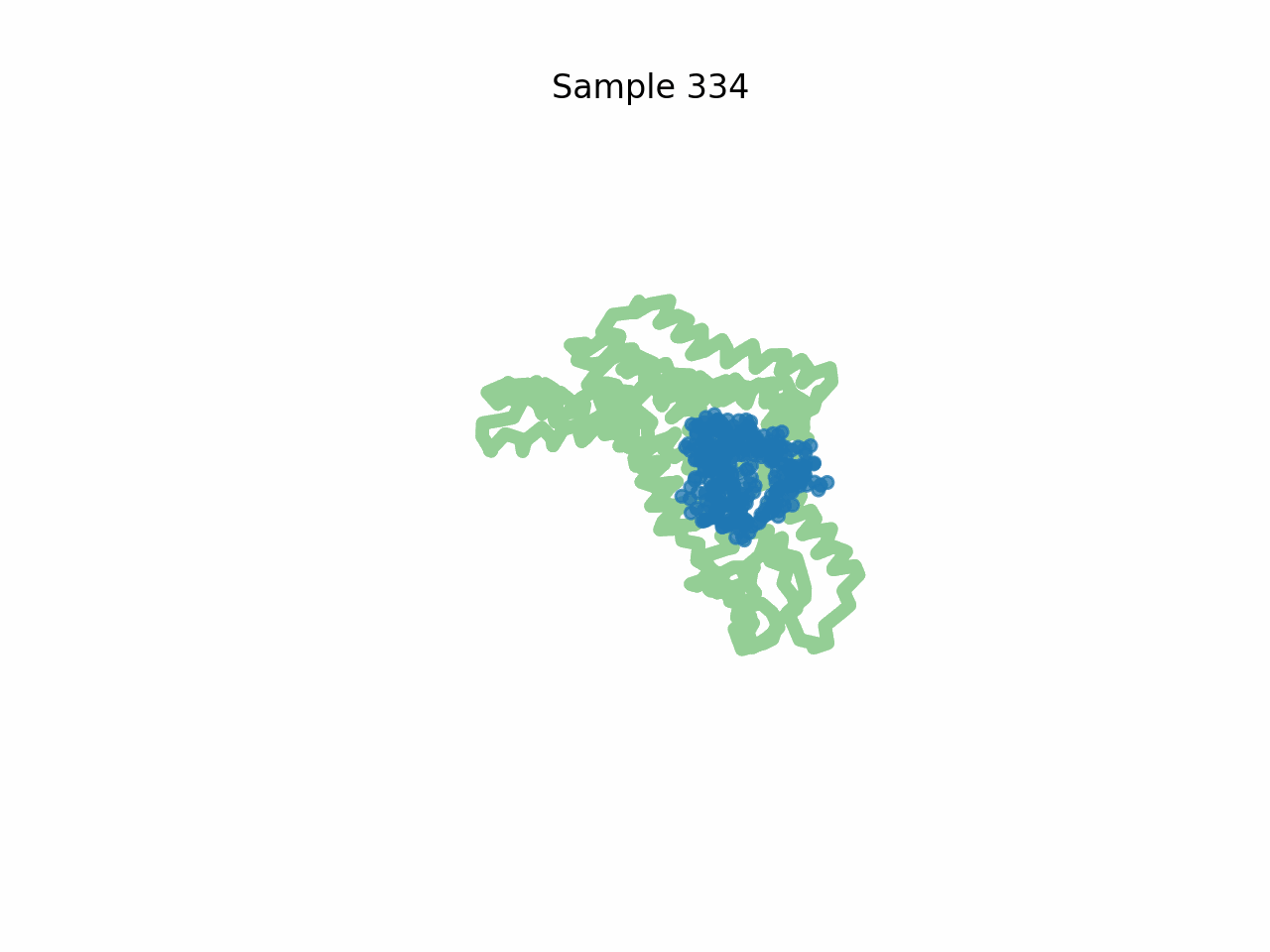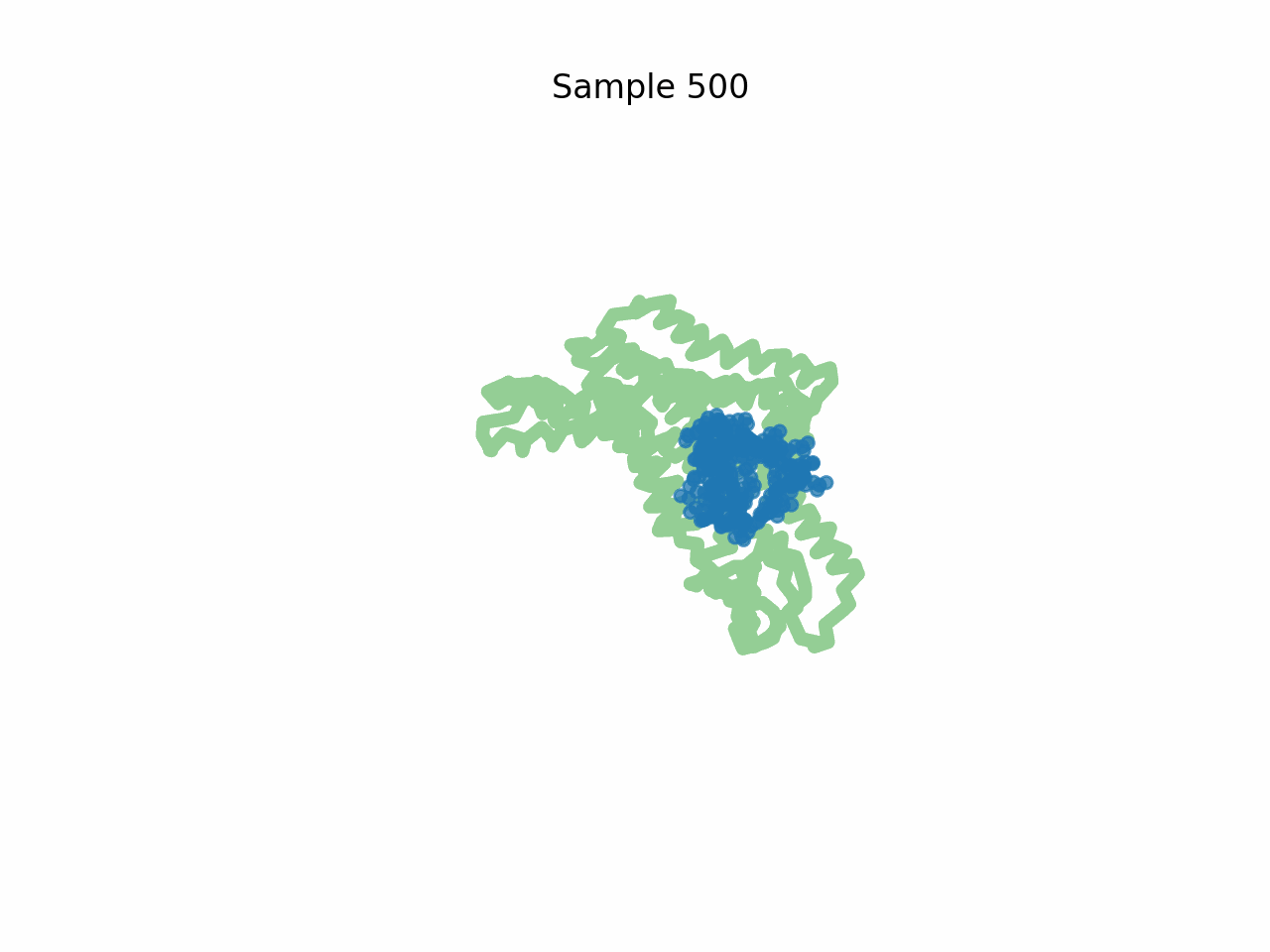
Inhibitor design
Daumiller D, Giammarino F, Li Q, Sonnerborg A, Cena-Diez R, Bryant P. Single-Shot Design of a Cyclic Peptide Inhibitor of HIV-1 Membrane Fusion with EvoBind. bioRxiv. 2025. p. 2025.04.30.651413. doi:10.1101/2025.04.30.651413 Paper Code

Binder design
Li, Q., Vlachos, E.N. & Bryant, P. Design of linear and cyclic peptide binders from protein sequence information. Commun Chem 8, 211 (2025). https://doi.org/10.1038/s42004-025-01601-3 Paper Code
Bryant P., Elofsson A. Peptide binder design with inverse folding and protein structure prediction. Communications Chemistry. (2023); 6: 229. Paper
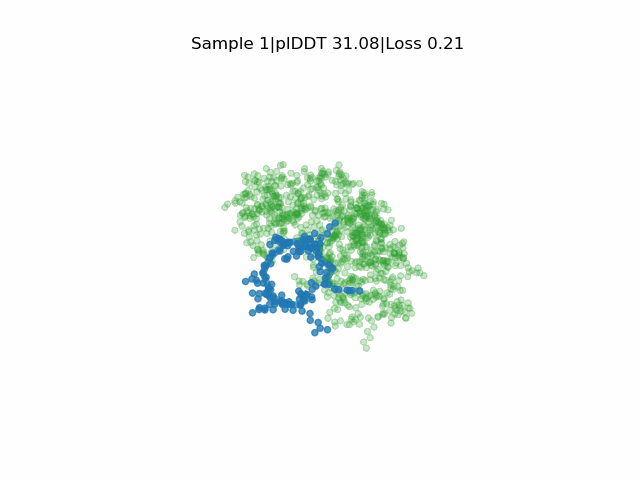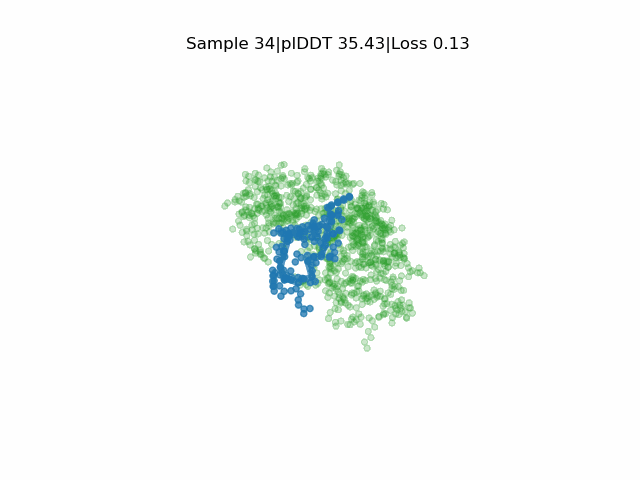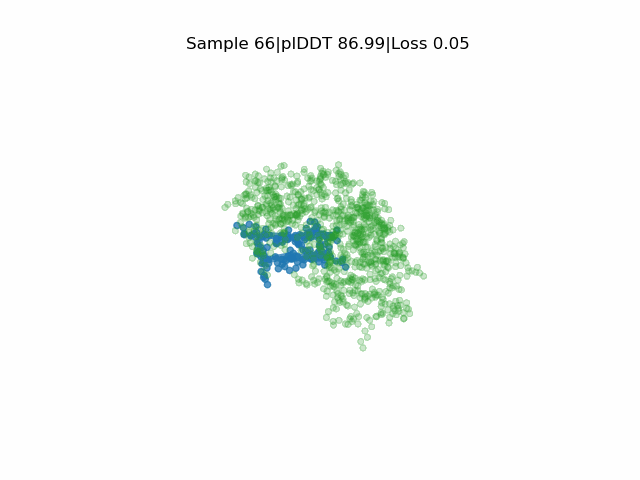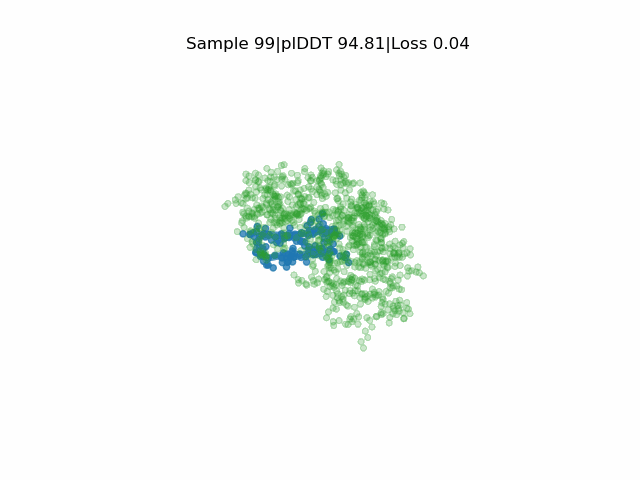
Structure prediction of protein-ligand complexes
Bryant, P., Kelkar, A., Guljas, A. Clementi, C. and Noé F. Structure prediction of protein-ligand complexes from sequence information with Umol. Nat Commun 15, 4536 (2024). https://doi.org/10.1038/s41467-024-48837-6 Paper Code
Structure prediction of alternative protein conformations
Bryant P, Noé F. Structure prediction of alternative protein conformations. Nat Commun 15, 7328 (2024). Paper Code

Structure prediction of protein complexes
Saluri M, Landreh M, Bryant P (2025) AI-first structural identification of pathogenic protein target interfaces. PLoS Comput Biol 21(6): e1013168. https://doi.org/10.1371/journal.pcbi.1013168 Paper Code
Bryant P, Noé F. Improved protein complex prediction with AlphaFold-multimer by denoising the MSA profile. PLoS Comput Biol. (2024);20: e1012253. Paper Code
Bryant, P., Pozzati, G., Zhu, W. et al. Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search. Nat Commun 13, 6028 (2022). Paper Code
Bryant P., Pozzati G., Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 13: 1–11 (2022). Paper Code Better code
Burke, D.F.^, Bryant, P.^, Barrio-Hernandez, I.^ et al. Towards a structurally resolved human protein interaction network. Nat Struct Mol Biol (2023). https://doi.org/10.1038/s41594-022-00910-8 Paper